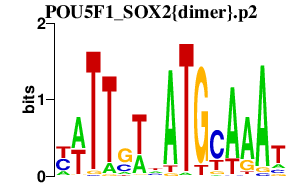
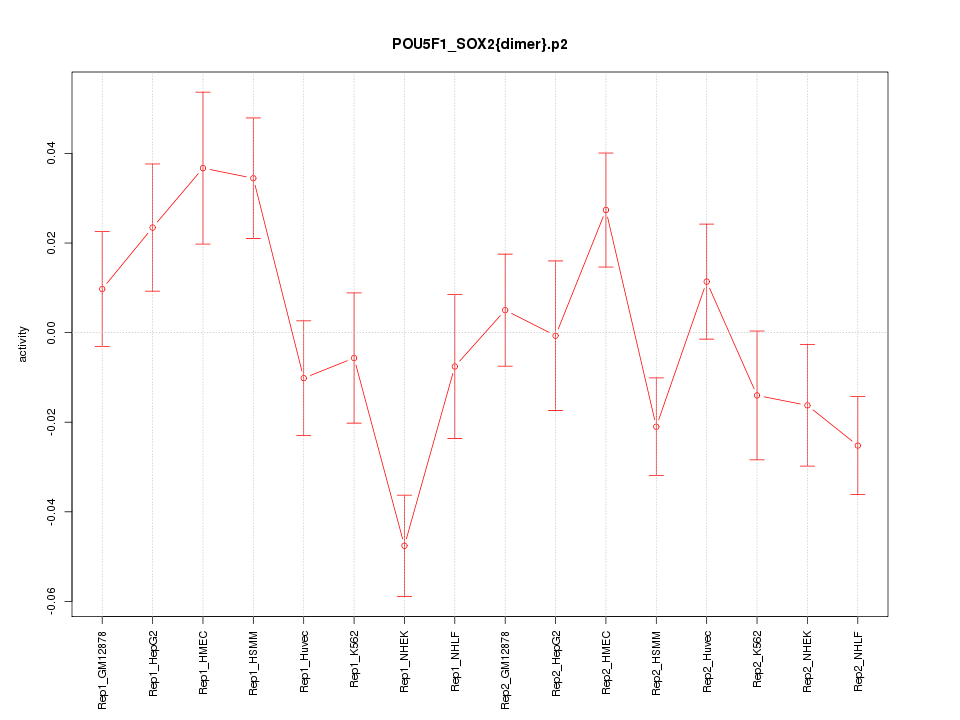
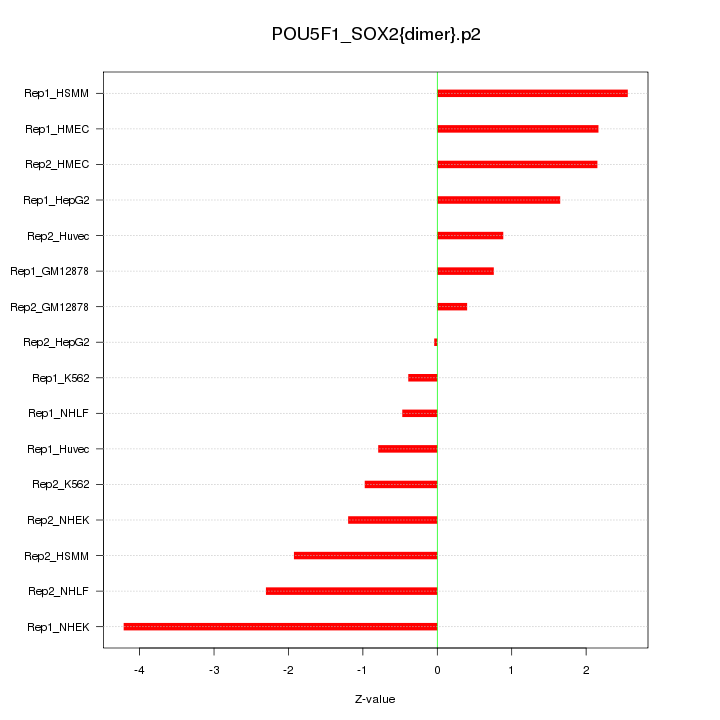
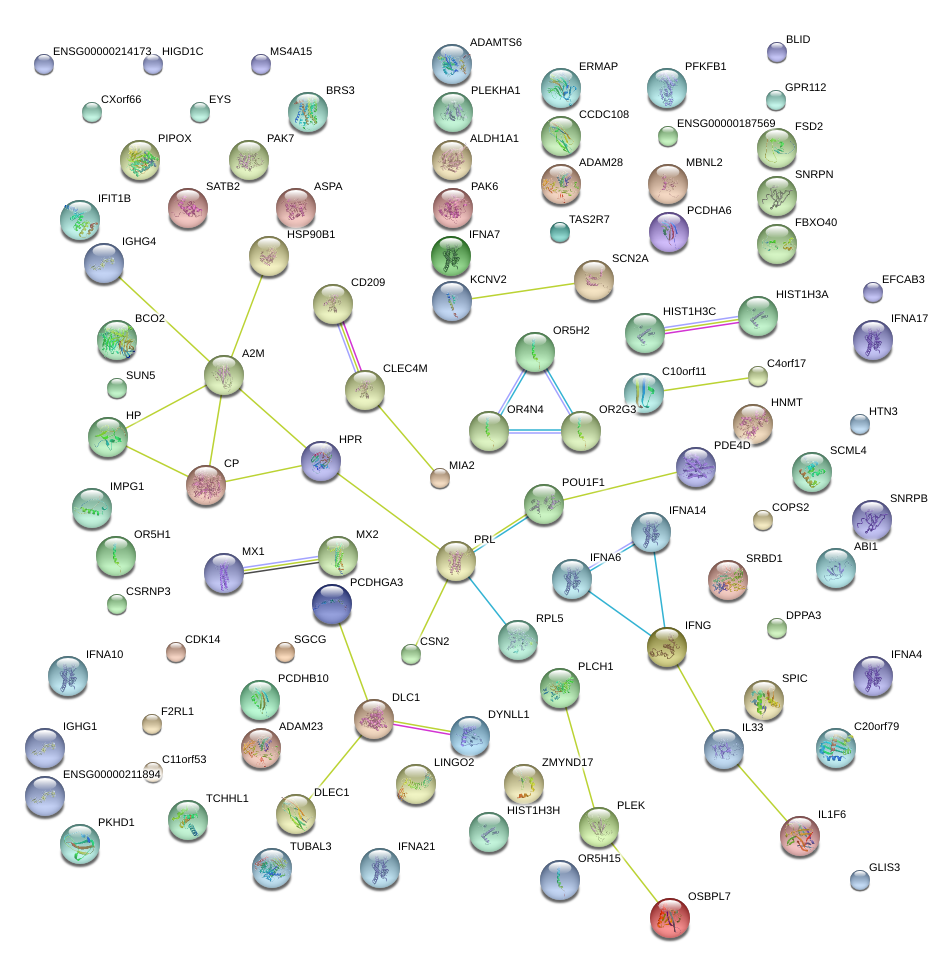
|
chr13_+_96592051
|
3.326
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_+_90176647
|
2.094
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr20_+_18742369
|
1.728
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr17_+_3326045
|
1.506
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr1_-_150328153
|
1.451
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr2_-_45648608
|
1.438
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr12_+_102859354
|
1.375
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr17_+_57811645
|
1.343
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr12_-_66839589
|
1.319
|
NM_000619
|
IFNG
|
interferon, gamma
|
|
chr6_-_66346417
|
1.298
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr2_+_166034402
|
1.214
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr1_+_93072947
|
1.172
|
|
RPL5
|
ribosomal protein L5
|
|
chr12_-_10846435
|
1.081
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr10_+_91127792
|
1.061
|
NM_001010987
|
IFIT1B
|
interferon-induced protein with tetratricopeptide repeats 1B
|
|
chr1_+_245835478
|
1.055
|
NM_001001914
|
OR2G3
|
olfactory receptor, family 2, subfamily G, member 3
|
|
chr4_-_151993422
|
1.017
|
|
|
|
|
chr14_-_106154332
|
1.007
|
|
LOC100126583
IGHG1
|
hypothetical LOC100126583
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_+_24394281
|
1.000
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr9_+_6205804
|
0.976
|
|
IL33
|
interleukin 33
|
|
chrX_+_135397790
|
0.944
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr9_-_21340885
|
0.936
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr9_-_21197141
|
0.931
|
NM_002171
|
IFNA10
|
interferon, alpha 10
|
|
chr15_-_81271859
|
0.905
|
NM_001007122
|
FSD2
|
fibronectin type III and SPRY domain containing 2
|
|
chr9_+_2707525
|
0.903
|
NM_133497
|
KCNV2
|
potassium channel, subfamily V, member 2
|
|
chr4_-_70861301
|
0.901
|
NM_001891
|
CSN2
|
casein beta
|
|
chr21_+_41663853
|
0.890
|
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr3_+_99484421
|
0.870
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chr9_-_21229975
|
0.865
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr9_-_4142182
|
0.862
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_+_165858586
|
0.859
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr10_-_5436781
|
0.854
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr17_+_24394043
|
0.842
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr11_+_111551417
|
0.842
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr11_+_110631916
|
0.821
|
NM_198498
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr12_+_49634048
|
0.821
|
NM_001109619
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C
|
|
chr15_+_19883745
|
0.818
|
NM_001005241
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4
|
|
chr2_-_219614481
|
0.809
|
NM_152389
NM_194302
|
CCDC108
|
coiled-coil domain containing 108
|
|
chr20_-_31055892
|
0.796
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr2_+_138438277
|
0.790
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr6_-_76838944
|
0.767
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr9_-_74757735
|
0.765
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr5_+_27508139
|
0.759
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr5_-_64813459
|
0.755
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr12_-_9123126
|
0.745
|
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_+_135210787
|
0.738
|
NM_153834
|
GPR112
|
G protein-coupled receptor 112
|
|
chr7_+_37689903
|
0.733
|
|
|
|
|
chr2_+_113479919
|
0.730
|
NM_014440
|
IL1F6
|
interleukin 1 family, member 6 (epsilon)
|
|
chr9_-_28709302
|
0.723
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr15_+_38296920
|
0.721
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr5_+_140751666
|
0.710
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr10_+_77212524
|
0.707
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr5_+_76150637
|
0.689
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chrX_-_55037235
|
0.684
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr10_-_27097782
|
0.682
|
|
ABI1
|
abl-interactor 1
|
|
chr14_+_38772870
|
0.680
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr11_+_60280915
|
0.672
|
NM_001098835
NM_152717
|
MS4A15
|
membrane-spanning 4-domains, subfamily A, member 15
|
|
chr2_+_207115060
|
0.670
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr6_-_22410993
|
0.669
|
NM_001163558
|
PRL
|
prolactin
|
|
chr1_-_93172119
|
0.656
|
|
|
|
|
chr5_+_140552087
|
0.652
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr12_+_7755316
|
0.647
|
NM_199286
|
DPPA3
STELLAR
|
developmental pluripotency associated 3
developmental pluripotency associated 3 pseudogene
|
|
chr11_-_121492010
|
0.629
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr3_-_156904690
|
0.627
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr4_+_100651183
|
0.625
|
NM_032149
|
C4orf17
|
chromosome 4 open reading frame 17
|
|
chr3_+_122794859
|
0.624
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr10_+_124141808
|
0.618
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr10_-_74863303
|
0.616
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr20_-_9767406
|
0.615
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr6_-_52060327
|
0.614
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr15_+_22619886
|
0.595
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr3_-_87408301
|
0.587
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr17_+_3324096
|
0.586
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr2_-_138762909
|
0.584
|
|
|
|
|
chr5_-_58331515
|
0.583
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_43063817
|
0.581
|
NM_018538
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chr19_+_7734034
|
0.577
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr15_+_22652790
|
0.575
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chrX_-_138875342
|
0.573
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr2_+_68445821
|
0.572
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr4_+_70928718
|
0.572
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr3_-_150422474
|
0.571
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr2_-_200038055
|
0.567
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr17_-_43254132
|
0.563
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr3_+_99334230
|
0.560
|
NM_001005338
|
OR5H1
|
olfactory receptor, family 5, subfamily H, member 1
|
|
chr6_-_108252200
|
0.551
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr5_+_140235114
|
0.548
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr6_+_26153590
|
0.547
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr13_+_22653059
|
0.543
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr16_+_70646008
|
0.540
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr18_+_74957475
|
0.534
|
|
|
|
|
chr3_-_106903734
|
0.534
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_-_41398126
|
0.531
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr5_+_140532385
|
0.528
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr5_+_140719773
|
0.522
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr18_-_73834670
|
0.521
|
|
|
|
|
chr17_-_43254117
|
0.519
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr11_-_35398077
|
0.516
|
NM_001195728
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr10_+_96512447
|
0.514
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr7_+_129057152
|
0.514
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr6_-_43384507
|
0.513
|
NM_206922
|
CRIP3
|
cysteine-rich protein 3
|
|
chr6_+_123142559
|
0.510
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr4_+_113437947
|
0.509
|
NM_001102406
NM_025144
|
ALPK1
|
alpha-kinase 1
|
|
chr3_+_28406990
|
0.507
|
NM_001040432
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chr12_-_90097384
|
0.505
|
NM_133503
|
DCN
|
decorin
|
|
chr7_+_29486010
|
0.505
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr7_-_36730525
|
0.502
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr5_+_76150588
|
0.498
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr1_+_69998445
|
0.489
|
NM_020794
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr2_-_233349469
|
0.483
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr14_-_106330781
|
0.479
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 pseudogene
|
|
chr1_+_156842852
|
0.479
|
NM_001004478
|
OR10Z1
|
olfactory receptor, family 10, subfamily Z, member 1
|
|
chr4_-_21155376
|
0.475
|
NM_001035004
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr1_-_150148608
|
0.472
|
NM_053055
|
THEM4
|
thioesterase superfamily member 4
|
|
chr13_-_109600709
|
0.471
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr6_+_64344307
|
0.468
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr11_-_128242342
|
0.466
|
NM_153764
NM_153765
NM_153766
NM_153767
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr9_-_99740243
|
0.466
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr1_-_100003936
|
0.466
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr20_+_16312
|
0.465
|
NM_153325
|
DEFB125
|
defensin, beta 125
|
|
chr2_+_191009265
|
0.464
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr15_+_63124760
|
0.463
|
NM_178859
|
OSTBETA
|
organic solute transporter beta
|
|
chr7_-_16811131
|
0.458
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr5_+_173405343
|
0.456
|
|
HMP19
|
HMP19 protein
|
|
chr6_+_32515596
|
0.451
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr7_-_84589182
|
0.449
|
NM_152754
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr9_+_104797413
|
0.448
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr2_-_165768798
|
0.448
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr5_+_173405298
|
0.444
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr7_-_122422989
|
0.438
|
NM_016945
|
TAS2R16
|
taste receptor, type 2, member 16
|
|
chr3_+_51837608
|
0.436
|
NM_001085479
|
IQCF3
|
IQ motif containing F3
|
|
chr5_-_24535386
|
0.434
|
NM_001190450
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr13_-_27441143
|
0.433
|
|
CDX2
|
caudal type homeobox 2
|
|
chr9_-_36365965
|
0.433
|
|
RNF38
|
ring finger protein 38
|
|
chr21_-_30583702
|
0.429
|
NM_001128598
|
KRTAP25-1
|
keratin associated protein 25-1
|
|
chr12_-_119733062
|
0.427
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr7_+_89681069
|
0.424
|
NM_001040666
|
STEAP2
|
six transmembrane epithelial antigen of the prostate 2
|
|
chr11_-_57964221
|
0.423
|
NM_001004733
|
OR5B12
|
olfactory receptor, family 5, subfamily B, member 12
|
|
chr3_-_38810504
|
0.420
|
NM_006514
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit
|
|
chr1_+_168381811
|
0.419
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr18_+_59571256
|
0.415
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr6_-_88917600
|
0.412
|
NM_001160260
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr10_+_95838801
|
0.410
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr12_+_103221590
|
0.408
|
NM_001008394
|
EID3
|
EP300 interacting inhibitor of differentiation 3
|
|
chr6_-_167491305
|
0.408
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr11_-_62508946
|
0.407
|
NM_004790
NM_153276
NM_153277
NM_153278
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6
|
|
chr11_-_59390518
|
0.407
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chrX_+_79562601
|
0.405
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr3_+_133798770
|
0.405
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr10_-_32707549
|
0.404
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr14_-_106149623
|
0.404
|
|
|
|
|
chr5_-_43448174
|
0.404
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr5_+_140227951
|
0.404
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr3_-_153541468
|
0.400
|
NM_001123228
|
TMEM14E
|
transmembrane protein 14E
|
|
chr6_-_119512035
|
0.400
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr5_+_140541163
|
0.399
|
NM_020957
|
PCDHB16
|
protocadherin beta 16
|
|
chr8_-_11891168
|
0.393
|
NM_001033019
|
DEFB134
|
defensin, beta 134
|
|
chr12_+_54106304
|
0.393
|
NM_001005183
|
OR6C76
|
olfactory receptor, family 6, subfamily C, member 76
|
|
chr2_-_171631728
|
0.392
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr1_+_57093030
|
0.389
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr1_-_229071957
|
0.384
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr4_+_70831387
|
0.381
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chr10_-_116408046
|
0.378
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_+_59804634
|
0.378
|
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chr4_+_185063421
|
0.377
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr5_-_135318420
|
0.376
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr5_+_140241976
|
0.375
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr1_-_151296611
|
0.374
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr11_+_82078217
|
0.374
|
|
|
|
|
chr5_+_79366746
|
0.374
|
NM_003248
|
THBS4
|
thrombospondin 4
|
|
chr19_-_62370667
|
0.373
|
NM_001012729
|
DUXA
|
double homeobox A
|
|
chr13_+_75092570
|
0.371
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr18_+_61568825
|
0.365
|
|
CDH7
|
cadherin 7, type 2
|
|
chr14_-_76958984
|
0.365
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr11_-_9182356
|
0.363
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr12_-_69600514
|
0.363
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr14_+_87560646
|
0.362
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr14_+_38720085
|
0.360
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr6_-_50045296
|
0.360
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chrX_+_70255128
|
0.360
|
NM_005120
|
MED12
|
mediator complex subunit 12
|
|
chr14_+_31868229
|
0.359
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr8_-_120720200
|
0.356
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr12_-_14887648
|
0.356
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr10_-_98109048
|
0.353
|
NM_001040102
NM_001040103
NM_033207
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein
|
|
chr21_-_35174742
|
0.353
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_-_21192201
|
0.348
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr21_-_30884586
|
0.347
|
NM_001164434
|
KRTAP22-2
|
keratin associated protein 22-2
|
|
chr4_+_120276386
|
0.346
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr6_+_131190237
|
0.345
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr12_+_7833292
|
0.343
|
|
NANOG
|
Nanog homeobox
|
|
chr3_+_2908920
|
0.342
|
NM_175613
|
CNTN4
|
contactin 4
|
|
chr7_-_122126422
|
0.340
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr9_+_21399132
|
0.339
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr2_+_211166323
|
0.339
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr4_+_77575276
|
0.337
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr8_-_7710547
|
0.336
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr8_+_42671718
|
0.335
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr6_+_132000134
|
0.335
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr8_+_32905027
|
0.334
|
|
|
|
|
chr4_-_152368492
|
0.334
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr17_+_18700385
|
0.332
|
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|